1.3 DNA polymorphism
1.3.1 Restriction Fragment Length Polymorphisms (RFLP)
RLFP are detected using a certain type of enzymes that cut DNA (restriction enzymes) at specific restriction site. Some restriction sites are polymorphic with one allele displaying the correct sequence for the restriction site while the second allele have an altered sequence so the restriction site is no longer recognized by the enzyme. The consequence is that the two adjacent restriction fragments remain linked together after treatment with the enzyme, leading to a polymorphism known as RFLP. The RFLP markers can be detected using molecular biology techniques such as southern hybridization or polymerase chain reaction (PCR) (see (Garibyan and Avashia 2013) for more detail).
1.3.2 Simple Sequence Length Polymorphisms (SSLP)
SSLP are repeated nucleotidic sequences displaying different numbers of repeat units in each allele. There are two types of SSLP: minisatellites with repeat unit up to 25 base pair3 (bp) in length and microsatellites with shorter repeated sequences (13 bp or less). Microsatellites are more commonly used than minisatellites because they are more frequent and evenly spread on the genome (\(5 \times 10^5\) with repeat units of 6 bp or less in the human genome). Furthermore, the PCR used to type a length polymorphism is more efficient and accurate with sequences less than 300 bp in length.
1.3.3 Single Nucleotide Polymorphisms (SNP)
A single nucleotide polymorphism is a variation in a single nucleotide that occurs at a specific position in the genome (see Figure 1.5). In a given population, most individuals may have a specific nucleotide at one position (e.g., a C) but a minority of individuals could have a different nucleotide at the same position (e.g., a G). The two possible nucleotide variations at a particular genomic position (locus) are said to be alleles, this type of polymorphism is extremely frequent in the human genome (a few millions).
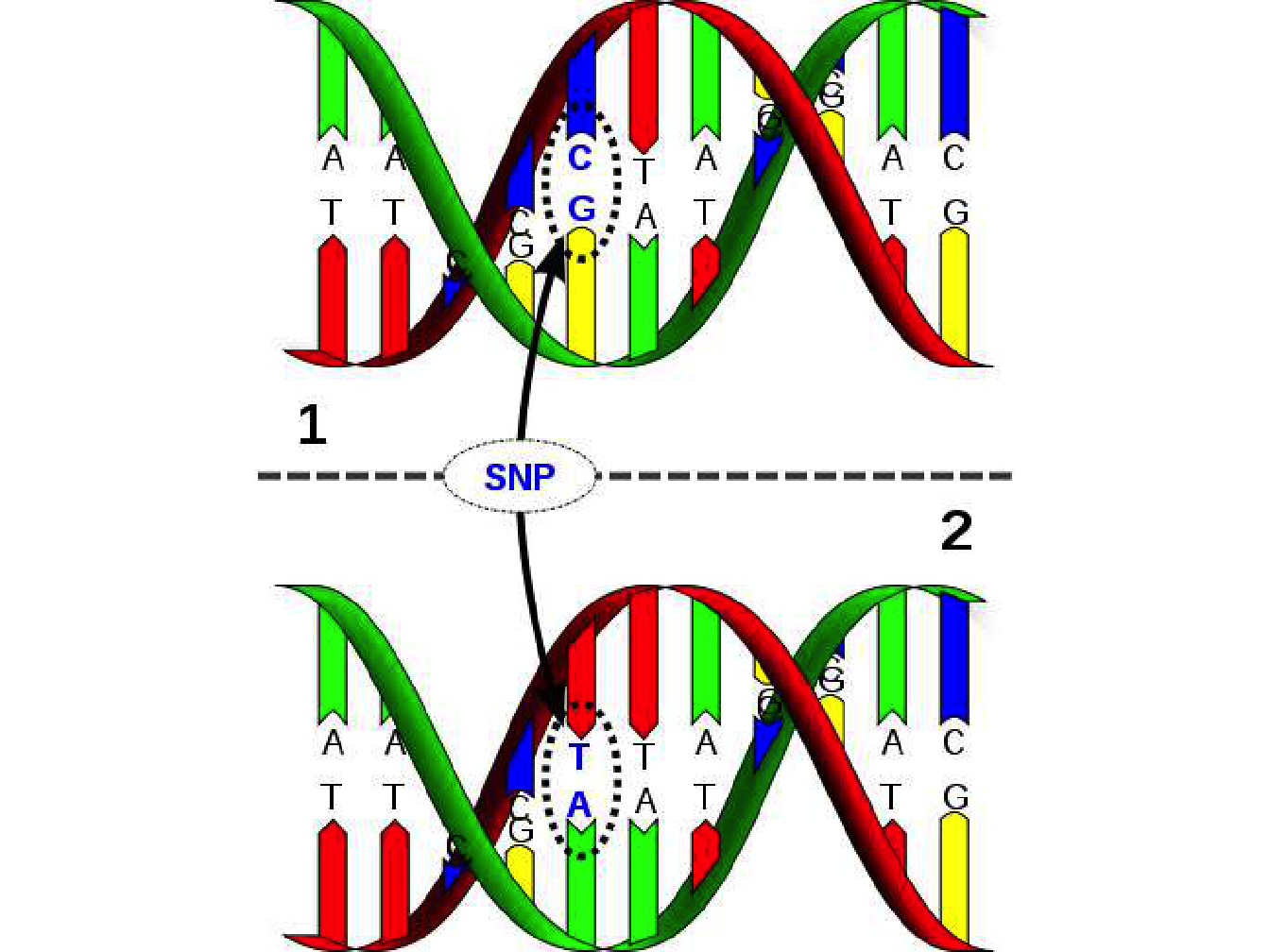
Figure 1.5: Schematic representation of a single nucleotide polymorphism. David Hall / Licence
The vast majority of SNP are biallelic because they originate from a point mutation in the genome, converting a nucleotide into another. For an SNP to be more than biallelic, it would be necessary for a new mutation to appear, after the first has been fixed in the population, to exactly the same position in the genome, which is highly unlikely. SNP typing methods are based on oligonucleotide hybridization analysis where an oligonucleotide (short single-stranded DNA molecule) will hybridize with another DNA molecule only if the oligonucleotide forms a completely base-paired structure with the other molecule (under precise temperature conditions).
Oligonucleotide hybridization can discriminate between the two alleles of an SNP if there is at least one mismatch at one position between the oligonucleotide and the target DNA. Several screening methods based on oligonucleotide hybridization exists: DNA chip (microarray) which use fluorescent markers to detect hybridization, oligonucleotide ligation assay (OLA) using capillary electrophoresis and amplification refractory mutation system (ARMS test) based on PCR primers and electrophoresis.
Recent breakthroughs in microarray technology have meant that hundreds of thousands of SNP can now be densely genotyped at moderate cost. As a result, it has become possible to characterize the genome of an individual with up to a million genetic markers. DNA chip technology makes use of piece of glass, or silicon, carrying many different oligonucleotides in a high-density array (Figure 1.6). To prepare really high-density arrays, oligonucleotides are synthesized in situ on the surface of the piece of glass resulting in a DNA chip. A density of up to 300,000 oligonucleotides per \(cm^2\) is possible and 150,000 polymorphisms can be typed in a single experiment (Brown et al. 2007).
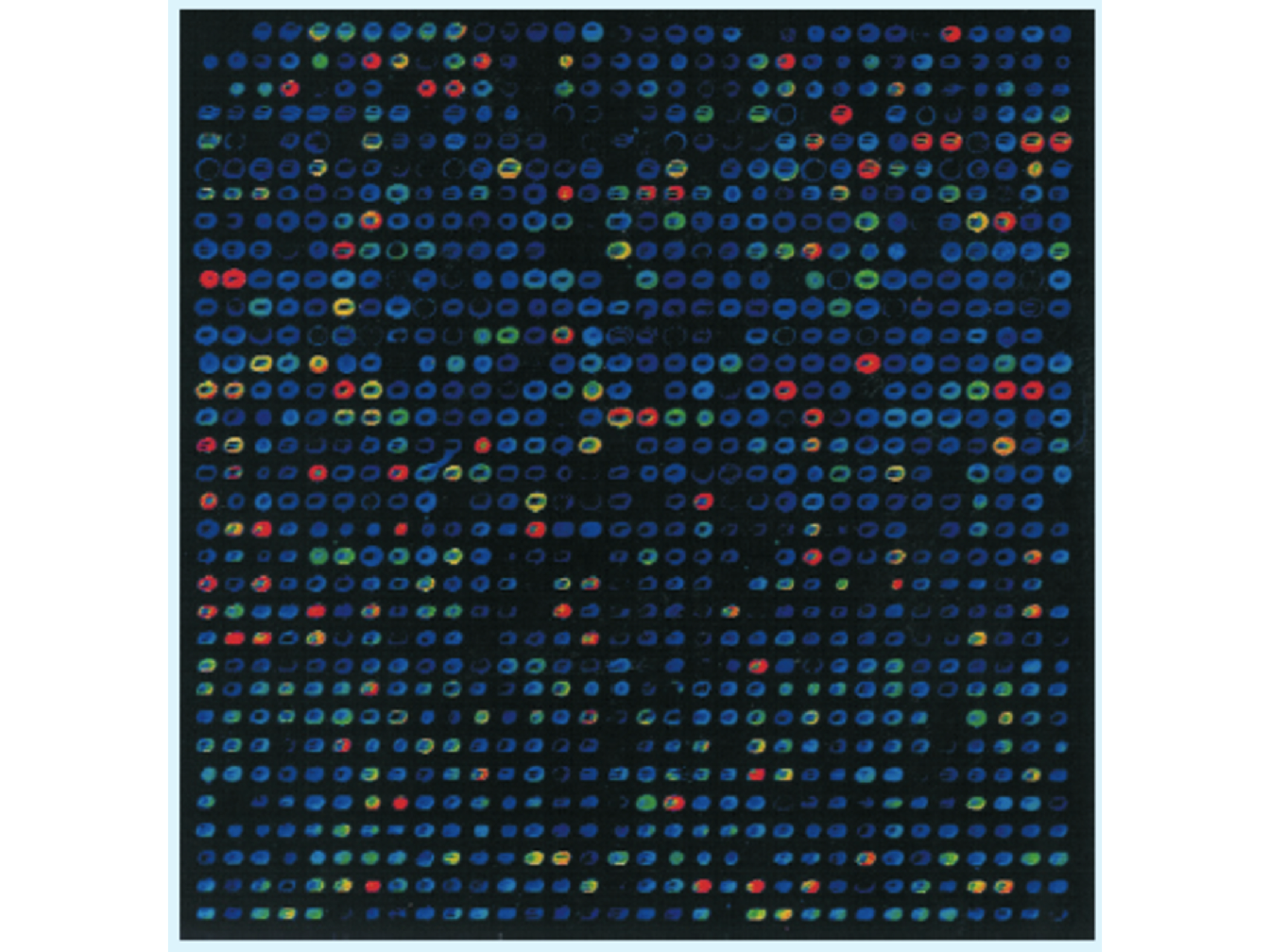
Figure 1.6: Visualization of the hybridization of a fluorescent labelled probe to a microarray. The DNA to be tested is labelled with a fluorescent marker and put onto the surface of the microarray. Hybridization is detected by examining with a fluorescence microscope the position at which the fluorescent signal is emitted indicating which oligonucleotides have hybridized with the target DNA (Brown et al. 2007).
Today, DNA microarrays are used in clinical diagnostic tests for some diseases. With the advent of new DNA sequencing technologies, some of the tests for which microarrays were used in the past now use DNA sequencing instead. Nevertheless, microarray tests being less expensive than sequencing, they remain used for very large studies as well as for some clinical tests.
References
Brown, T.A., D.B.S.T. Brown, T.A. Brown, and L.B.T. Brown. 2007. Genomes 3. Taylor & Francis Group, an Informa Business. Garland Science Pub. https://books.google.fr/books?id=Cjl98tqp6rsC.
Garibyan, Lilit, and Nidhi Avashia. 2013. “Research Techniques Made Simple: Polymerase Chain Reaction (Pcr).” The Journal of Investigative Dermatology 133 (3): e6.
A base pair (bp) is a unit consisting of two nucleobases bound to each other by hydrogen bonds.↩